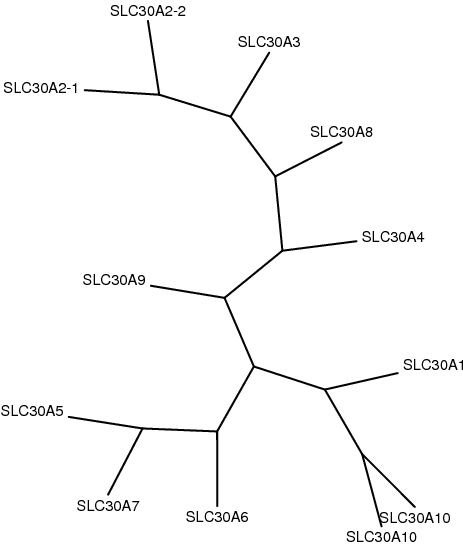
Phylogenetic trees were constructed using PhyML and drawn using Phylip, based on multiple sequence alignments by MUSCLE.
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
--- PhyML v3.0 (179M) ---
http://www.atgc-montpellier.fr/phyml
Copyright CNRS - Universite Montpellier II
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
. Sequence filename: slc30-muscle.fa.fas.phylip
. Data set: #1
. Tree topology search : Best of NNIs and SPRs
. Initial tree: BioNJ
. Model of amino acids substitution: LG
. Number of taxa: 12
. Log-likelihood: -21888.28877
. Unconstrained likelihood: -12557.36316
. Parsimony: 4150
. Tree size: 18.10003
. Discrete gamma model: Yes
- Number of categories: 4
- Gamma shape parameter: 1.753
. Time used 0h5m57s
. 357 seconds
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
Suggested citation:
S. Guindon & O. Gascuel
"A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood"
Systematic Biology. 2003. 52(5):696-704.
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo