About Pose & Rank
Pose & Rank is a free service for scoring protein-ligand complexes, using two atomic distance-dependent statistical scoring functions: PoseScore and RankScore [1]. PoseScore was optimized for recognizing native binding geometries of ligands from other poses. RankScore was optimized for distinguishing ligands from nonbinding molecules.
The input includes the 3D structure of the protein receptor, the binding/docking poses of small molecules in that protein. The output is a list of scores for each protein-small molecule complexes.
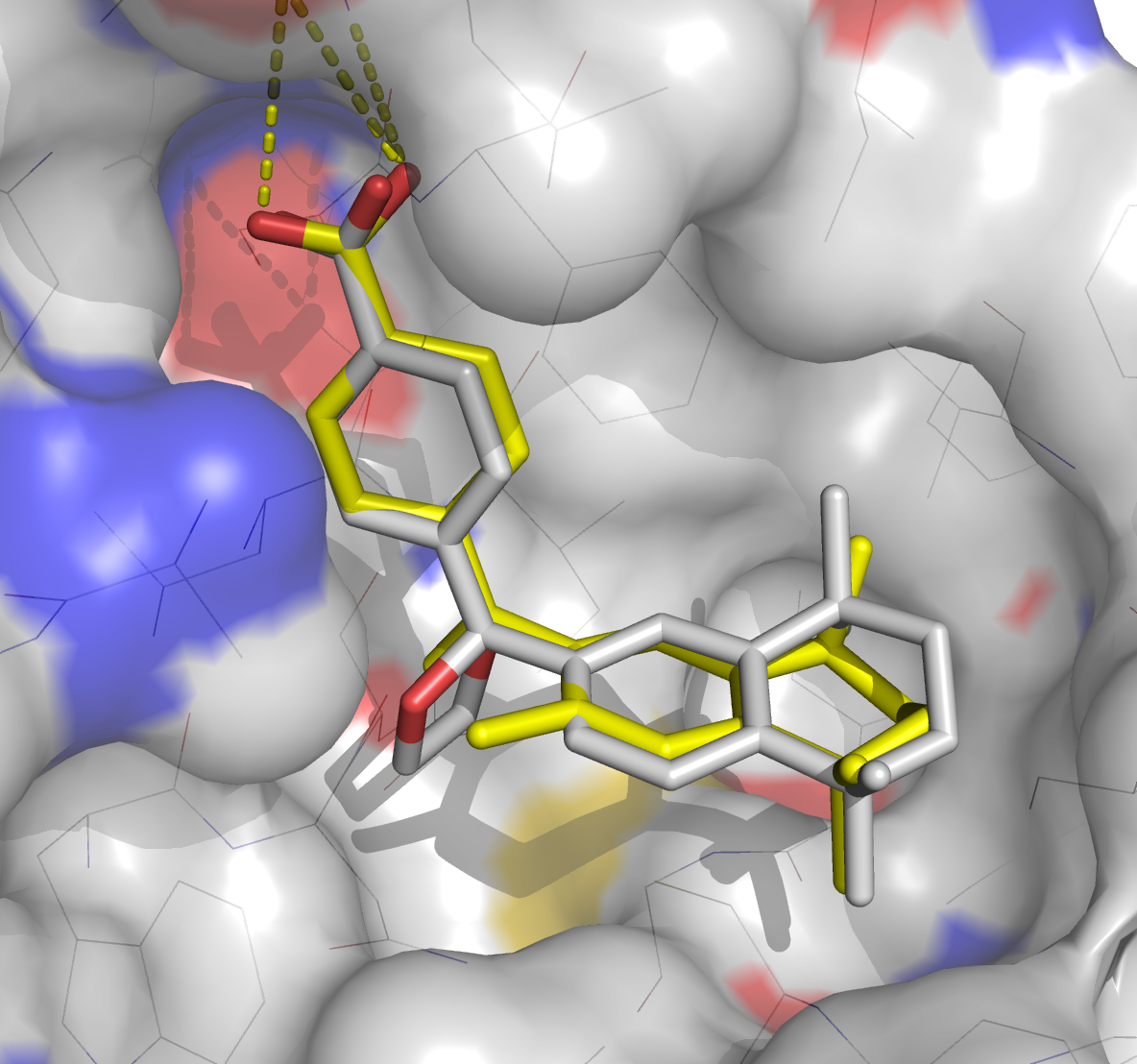
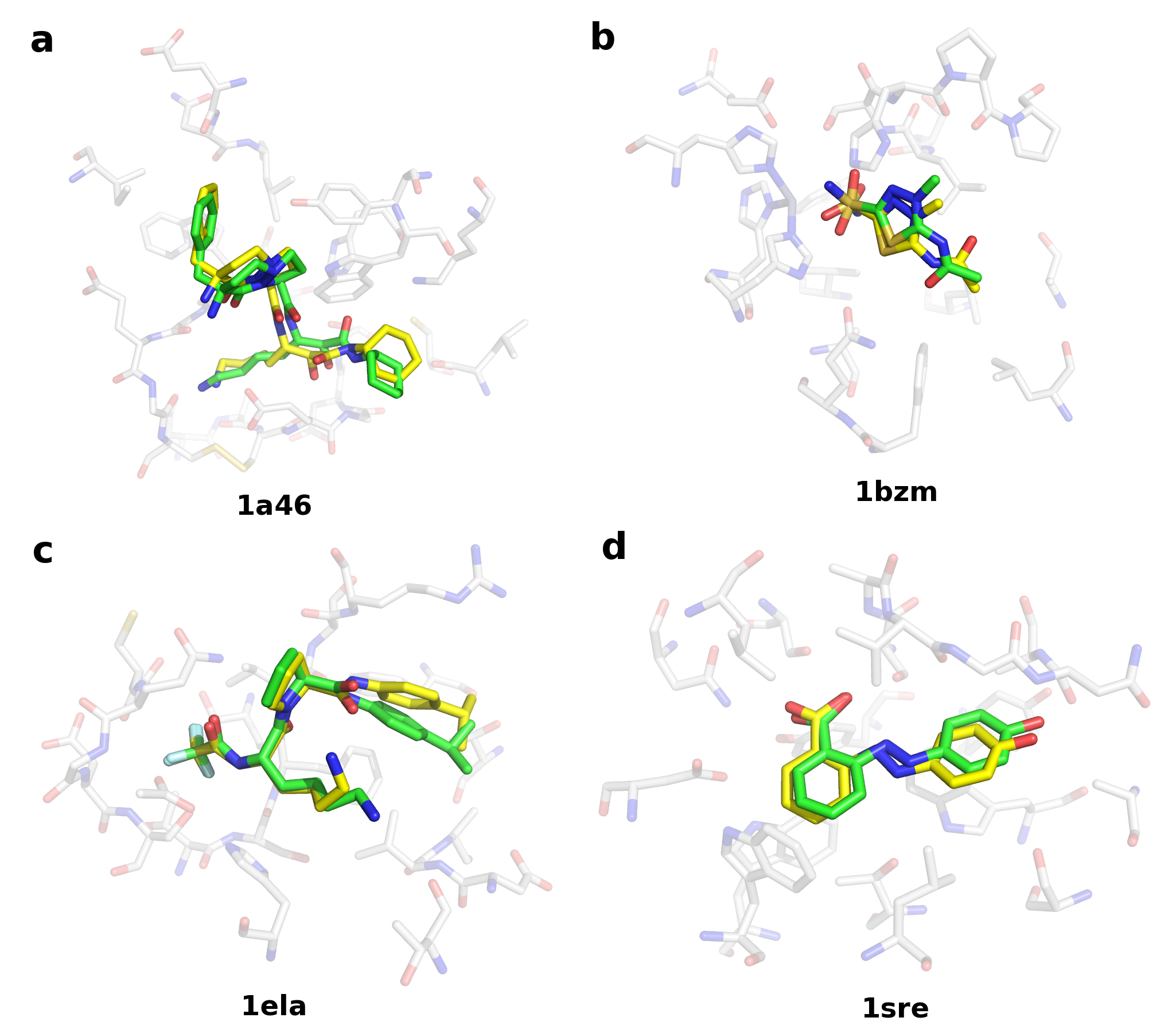
PoseScore ranks a native/near-native binding pose the best, top 5 and top 10 for 88%, 97% and 99% targets (Fig 1). PoseScore accuracy is comparable to that of DrugScore(CSD) and ITScore/SE, and superior to 12 other tested scoring functions. RankScore improved the ligand enrichment (logAUC) and early enrichment (EF1) scores computed by DOCK 3.6 for 68% and 74% targets, respectively. RankScore performed better at rescoring than each of seven other scoring functions tested. Therefore, PoseScore can be used for protein-ligand complex structure prediction, by ranking different conformations of a given protein-ligand pair. RankScore can facilitate ligand discovery, by ranking complexes of the target with different small molecules.
References
- Fan H, Schneidman D, Irwin JJ, Shoichet BK, Sali A. Statistical potential for modeling and ranking protein-ligand interactions. J. Chem. Inf. Model. 2011, 51:3078-92.
Fan H, Schneidman-Duhovny D, Irwin J, Dong GQ, Shoichet B, Sali A. Statistical Potential for Modeling and Ranking of Protein-Ligand Interactions. J Chem Inf Model. 2011, 51:3078-92. [ Abstract ]
Contact: