|  |
About FoXSDock
FoXSDock is a method for docking with SAXS profile of the complex. The input is two proteins and a SAXS profile of their complex. The output is a list of potential complexes sorted by a score that combines interface energy and the fit to SAXS profile.
Method Overview
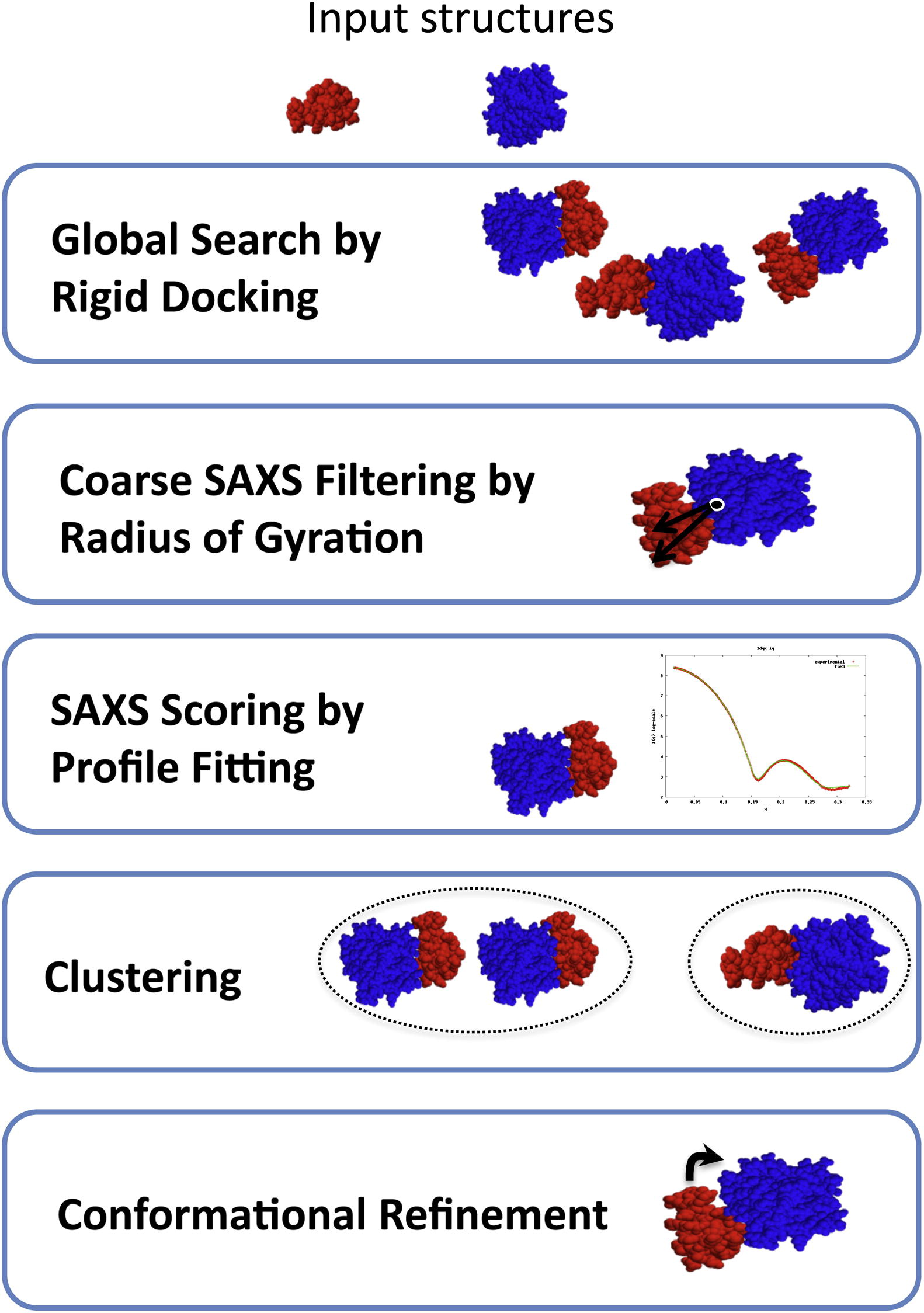
The docking protocol involves five steps (Fig. 1):- Global search. Rigid docking is performed by a geometric shape-matching algorithm PatchDock [1-2], generating thousands of models. In this step, the flexibility is taken into account implicitly by allowing a small amount of steric clashes at the interface.
- Coarse SAXS filtering. Radius of gyration predicted from the SAXS profile of the complex is used to filter out rigid docking models that do not agree with the SAXS measurements[3].
- SAXS scoring. Each docking model is fitted against the SAXS profile of the complex using FoXS method [4].
- Clustering. Remaining docking models are clustered by their interface Cα RMSD and the cluster representative with the best fit to the SAXS profile is selected.
- Conformational refinement. Cluster representatives are refined and steric clashes are removed through optimization of side chain positions and relative protein orientations, with FireDock [5]. The final models are scored and ranked by an energy-based score and the fit to the SAXS profile.
References
- Duhovny D, Nussinov R, Wolfson HJ. Efficient Unbound Docking of Rigid Molecules. In Gusfield et al., Ed. Proceedings of the 2'nd Workshop on Algorithms in Bioinformatics(WABI) Rome, Italy, Lecture Notes in Computer Science 2452, pp. 185-200, Springer Verlag, 2002 [ PDF file ]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. NAR 33: W363-367, 2005. [ Full text ]
- D. Schneidman-Duhovny, M. Hammel, and A. Sali.Macromolecular docking restrained by a small angle X-ray scattering profile. J Struct Biol. 2011. 173(3):461-71
- D. Schneidman-Duhovny, M. Hammel, and A. Sali. FoXS: A Web server for Rapid Computation and Fitting of SAXS Profiles. NAR 2010.38 Suppl:W540-4.
- N. Andrusier, R. Nussinov and H. J. Wolfson. FireDock: Fast Interaction Refinement in Molecular Docking.Proteins (2007), 69(1):139-159.
If you use FoXSDock (version main.73fcb34), please cite:
Schneidman-Duhovny D, Hammel M, Sali A. Macromolecular docking restrained by a small angle X-ray scattering profile. J Struct Biol. 2010 [ Abstract ]Schneidman-Duhovny D, Hammel M, Tainer JA, and Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. NAR 2016 [ FREE Full Text ]
Contact:

